A few weeks ago Kafkas et al. (2013) published a paper looking at current patterns of how datasets o biological databases are cited in research articles, based on an analysis of the full text Open Access articles available from Europe PMC. They identified data citations by:
- Accession numbers available in articles as publisher-supplied, structured content;
- Accession numbers identified in articles by text mining;
- References to articles from the ENA, UniProt and PDBe records.
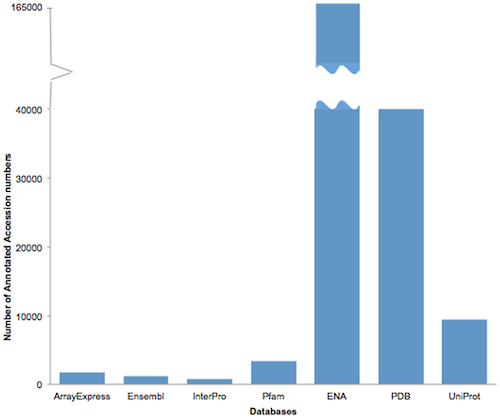
They could show that text mining doubles the number of structured annotations available in journal articles (from 2.26% to 5.15%), and that these structured annotations should be extended beyond the ENA, UniProt and PDB identifiers that their analysis focused on. ENA identifiers (for nucleotide sequences in GenBank, EMBL or DDBJ) make up the largest group, with 160,112 identifiers found in the 410,364 articles that were analyzed.
Another result in the paper is that references to articles in these databases show little overlap with database links found in articles. One of the conclusions drawn by the author is that
Text-mining can be used to extend structured data citation, and could be a basis for the development of services to help authors or editors to add structured content at the beginning of the publication process, rather than after the fact.
Adding structured data citations during the authoring phase of a manuscript requires tools that make this process easier, providing auto-linking and verification without requiring extra input from the author. Scholarly Markdown is an ideal platform for these tools, as it is easier to extend than traditional word processors such as Microsoft Word. During a small workshop around persistent identifiers for data (DataCite), people (ORCID) and geological samples (IGSN) that took place yesterday and today at the GFZ Potsdam I worked on a tool that does auto-linking for these identifiers:
- IGSN. International Geosample Number
- MGI identifiers for genetically modified mouse strains in the Internal Mouse Strain Resource
- ENA. Genbank / ENA / DDBJ nucleotide sequences
- UniProt protein sequences from the UniProt database
- PDB. Protein Data Bank protein structure information
The list includes the IGSN, the database identifiers studied by Kafkas et al (2013), and the MGI identifier for genetically altered mice. In the life sciences there is a long tradition - and requirement by journals - to use database identifiers for data, but identifiers for resources such as genetically modified mice are unfortunately not in common use.
This blog uses the Pandoc markdown processor and the Jekyll static website generator. The easiest way to implement this functionality was by writing a filter for the liquid templating engine used by Jekyll, and provide this filter as a Jekyll plugin. The Jekyll plugin can be found at mfenner/jekyll-scholmd. The plugin expects the name of the identifier, followed by a colon and optional space, followed by the identifier:
GenBank: M10090
IGSN: JRH964436
MGI: 96922
UniProt: P02144
PDB: 1mbnThis input is automatically translated into GenBank:M10090, IGSN:JRH964436, MGI:96922, and information about the human myoglobin protein (UniProt:P02144, PDB:1mbn) is generated in a similar fashion.
The plugin was written in a few hours today, and is my first Jekyll plugin. There is room for improvement, e.g. support for more identifiers, better regex matching, validation of the resulting links, and automated tag generation if an identifier is found. Ideally the auto-linking should happen in the markdown and not the HTML output, so that these structured database links are also available in other markdown outputs such as PDF. But this is another example how Scholarly Markdown can make it easier for researchers to author documents without requiring a fancy web-based user interface.
References
Kafkas, Ş., Kim, J.-H., & McEntyre, J. R. (2013). Database Citation in Full Text Biomedical Articles. PLoS ONE. doi:10.1371/journal.pone.0063184