Continuous Publishing
Earlier this week Björn Brembs wrote in a blog post (What Is The Difference Between Text, Data And Code?):
To sum it up: our intellectual output today manifests itself in code, data and text.
The post is about the importance of publication of data and software where currently the rewards are stacked disproportionately in favor of text publications. The intended audience is probably mainly other scientists (Björn is a neurobiologist) who are reluctant to publish data and/or code, but there is another interesting aspect to this.
Just as scientific publication increasingly means more than just text and includes data and software, we are also increasingly seeing tools and methodologies common in software development applied to scientific publishing. This in particular includes the ideas behind Open Source software (which shares many commonalities with Open Access and Open Science), but also tools like the git version control system (We Need a Github of Science) or the markdown markdown language (A Call for Scholarly Markdownjudgment).
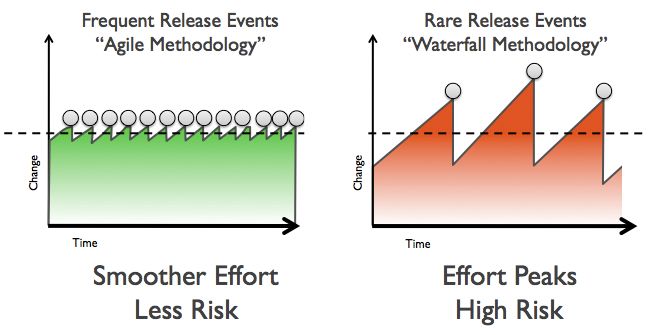
Continuous Delivery is another concept increasingly popular in software development that has many implications on how research can be performed and reported. Martin Fowler describes it as:
Continuous Delivery is a software development discipline where you build software in such a way that the software can be released to production at any time.
The concept of frequent small releases is of course familiar to everyone practicing Open Notebook Science, writing science blogs, presenting preliminary data at conferences or publishing preprints, and is even relevant to CrossMark, a service that tracks corrections, enhancements and other changes of scholarly documents.
When you read the definition given by Martin Fowler carefully, you see that Continuous Delivery is about more than the frequency of software updates – it is in fact about improving the process of releasing software. The scientific publication is the corresponding event in science, and I think that nobody would argue with me that the experience publishing a paper is too complex, time-consuming and often frustrating. The focus here is not on the time it takes to do peer review, or the multiple revisions needed before a manuscript is accepted. I am talking about the pain submitting a manuscript, the back and forth regarding file formats, citation styles and other technical requirements, the reformatting of manuscripts, and also the time it takes from accepting a manuscript to finally publishing it online.
I would argue that the main reason publishing is so painful for everyone involved is that it is still very much a manual process. Just as software development is creative work, but still can benefit tremendously from tools such as automated tests and build tools, we can apply the same principles to scientific publishing. This means that everything that can be automated should be automated so that we can focus on those areas that need human judgment. The mistake that I think is commonly made is that automation for many publishers means automation for the publisher, with even more work for the author who submits a manuscript. A good example is that authors are increasingly asked to submit publication-ready manuscripts even though typesetting and desktop publishing is not their area of expertise and the manuscript text will be very different after one or more rounds of revision. The pain of processing manuscripts into something that can be published was summarized perfectly by typesetter and friend Kaveh Bazargan at the SpotOn London 2012 Conference (via Ross Mounce’s blog):
It’s madness really. I’m here to say I shouldn’t be in business.
The promise of Continuous Delivery for publishing is to develop tools and best practices that make the process of publication faster, with better quality, and less frustrating. Continuous Integration (again Martin Fowler) is an important part of Continuous Delivery and means frequently merging all developer working copies of a software project into a central repository, combined with running automated unit tests and software builds using an integration server.
We can apply Continuous Integration to scholarly documents - instead of automated tests and software builds we can automate the transformation of documents into JATS XML and other output formats, and we can automate the process of checking for required metadata, correct file formats for images, etc. And we can use the same software tools for this, many of which are freely available to Open Source projects.
As an example of how this can be done I have integrated the Travis CI Continuous Integration server with the book project Opening Science. The recently published book is a dynamic book that hopefully is updated frequently in the coming months. Every time an editor approves a correction to the text - hosted in markdown format on Github - the Travis CI server is automatically triggered to build a new HTML version of the book and to push the new version to the book website. The Travis server is running the Pandoc document converter to not only convert the changed document from markdown to HTML, but Pandoc will also insert and format references, and the Jekyll site generator will build a nice website around the markdown files. Over time this build process can be extended to do other things as well, from auto-generating links to data and resources to transforming the document into other file formats besides HTML.
References
Brembs, B. (2014). What is the difference between text, data and code? https://doi.org/10.59350/1nnrk-reh85
Fenner, M. (2012). A Call for Scholarly Markdown. https://doi.org/10.53731/r294649-6f79289-8cw1g
Mounce, R. (2012). Yet another #solo12 recap (part2). https://doi.org/10.59350/hqrq0-6sc39
Fenner, M. (2013). From Markdown to JATS XML in one Step. https://doi.org/10.53731/r294649-6f79289-8cw0k
Fenner, M. (2013). Auto generating links to data and resources. https://doi.org/10.53731/r294649-6f79289-8cw16



Comments ()