One of the challenges of collecting metrics for scholarly outputs is persistent identifiers. For journal articles the Digital Object Identifier (DOI) has become the de-facto standard, other popular identifiers are the pmid from PubMed, the identifiers used by Scopus and Web of Science, and the arxiv ID for ArXiV preprints.
For other research outputs the picture is less clear. DOIs are also used for datasets, but so are many other identifiers, in particular in the life sciences.
To collect metrics for research outputs, the requirements are slightly different. We need identifiers understood by the services collecting the metrics, not by the data repository or other service that is holding the research output (the only exception is usage stats, which are generated locally). For many services, in particular social media such as Facebook, Twitter or Reddit, the primary identifier for a resource is a URL. This means that we should have one or more URLs for every research output where we want to track the metrics - typically the publisher or data repository landing page. Since URLs can be messy, Google, Facebook and others have come up with the concept of a canonical URL, and some care should go into constructing proper canonical URLs (see this blog post for examples of what can go wrong).
The Den Haag Manifesto is the result of a Knowledge Exchange workshop held in June 2011 and tries to bring Persistent Identifiers and Linked Open Data together. The first principle is very much in line with what I said above:
Make sure PIDs can be referred to as HTTP URI’s, including support for content negotiation.
Or, to put this differently: URLs are good enough to start collecting metrics for scholarly outputs. Scientific software is a good example where persistent identifiers are not commonly used (despite efforts such as this one), but we can still collect many meaningful metrics using the repository URL (and the open source software lagotto):
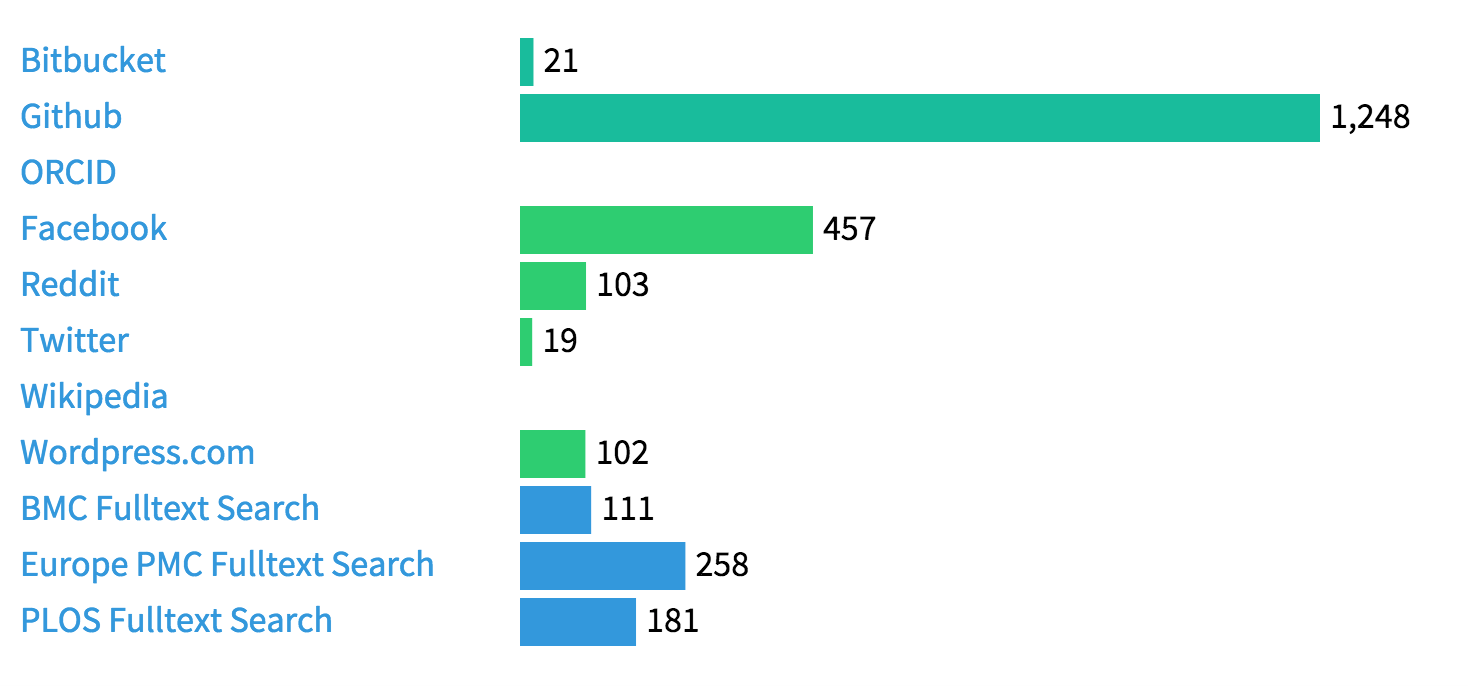
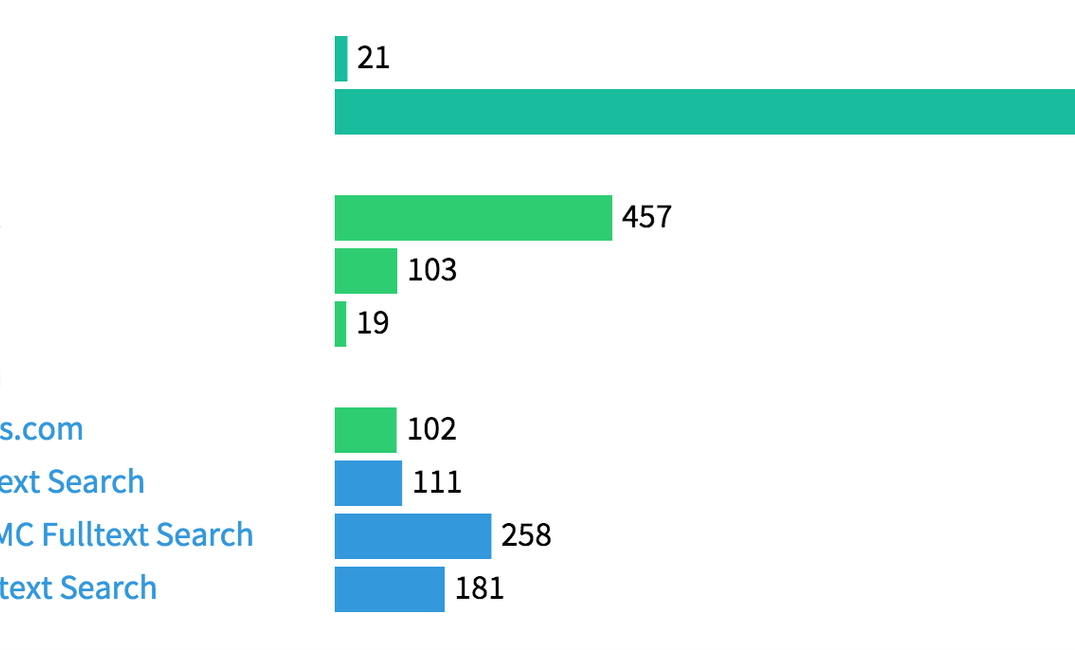
The last three rows are citations in the scholarly literature found via fulltext search of BioMed Central, Europe PMC and PLOS. URLs (in contrast to persistent identifiers represented as strings and/or numbers) are easy to find, the main limitation is not so much using a URL rather than a DOI, but that scientific software typically is mentioned in the text without appearing in the reference list. This makes it hard to impossible to find articles mentioning the software that are not open access, which unfortunately is still the majority of them.
We are of course also tracking the discussion of the software in social media, and are collecting the number of stars and forks in Github and Bitbucket. Overall there is quite a lot of activity, here are some examples:
- Windowed Adaptive Trimming for fastq files using quality
- Reads simulator
- Toolkit for processing sequences in FASTA/Q formats
All three software repos have been cited in the scholarly literature at least ten times. What is missing is infrastructure that tracks the citations of scientific software, so that we can give proper scientific credit to the authors of the software, and can discover other research projects using the same tools. software.lagotto.io uses a list of software repos collected by Jure Triglav for ScienceToolbox, and a scientific software index is indeed one of the important missing pieces.