In my last post I wrote about the importance of keeping things simple in scholarly publishing, today I want to go into more detail with one example: citations in scholarly documents.
Citations are an essential part of scholarly documents, and they are summarized in the references section at the end of the article or book chapter. The problem is that not everything that is cited in a scholarly document ends up in the references list. Examples of this include:
- web links, e.g. to reagents or other resources
- identifiers for biological databases such as GenBank that are typically included in the text as identifiers or as links
- footnotes with links to external resources
In other words: we are not consistent in how we cite other content. And this is a problem because we are making it more difficult than necessary for authors, publishers, and everyone else to handle these various citation flavors, and, more importantly, we are losing citations along the way. This is a particular problem for data citation, as the seminal 2013 paper by Kafkas et al. has shown for citations to the three biological databases ENA (European Nucleotide Archive), PDB, and Uniprot:
- there is a large number of accession numbers in the Open Access subset of PubMed Central (e.g. 160,112 ENA accession numbers for papers published up until June 2012)
- text mining using the Whatizit tool can retrieve most of these identifiers
- there is only partial overlap between database identifiers annotated by publishers and database identifiers found by text mining
- the overlap is even smaller between papers citing database identifiers, and papers cited in biological databases such as ENA
- the study was limited to Open Access journals, as only for them the full-text articles could be text mined
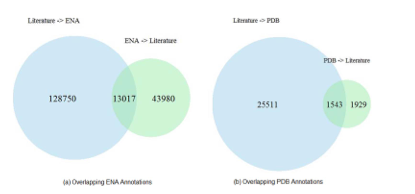
In other words, even though including identifiers for biological databases has been an accepted community standard that every author and publisher is following for a long time, the proper citation of these identifiers is still often broken. The picture doesn’t seem to be any better for DOIs for datasets: while they are fairly common by now, their use in scholarly articles differs widely from appearance in the references list to links in the materials and methods section to no mention at all.
There are various ways how this can be fixed (e.g. requiring authors to use biological database identifiers in a consistent way, better text mining tools, opening up subscription content to text mining), but the best solution is the simplest one: every citation in a paper should go into the references list. As an example I have added the ENA mRNA U65091 (Shioda, Fenner, & Isselbacher, 1997) - something I worked on a long time ago - to the references list of this post.
Technology
For this to work, it is essential that reference managers - the software authors use to generate the references list - properly support citations to data, including biological databases. It appears that all major reference managers support datasets as reference type and there is good community agreement what a data citation should look like (Joint Declaration of Data Citation Principles). What is missing is support for easily importing the required metadata for these datasets, and reference managers use two approaches for this:
- query external databases via API and pull in the required metadata (e.g. Papers, Endnote)
- browse to the webpage describing the database entry and import the metadata via bookmarklet/web importer (e.g. Zotero, Mendeley)
Both approaches require custom code for every database. Whereas many reference managers use Citation Style Language (CSL) as a standard way to format references, no such standard exists for web importers. Which means that every reference manager has to implement this separately, and most of them are not open source software so that the community could help.
PLOS Labs is holding a Citation Hackathon on October 18 in their San Francisco office. While I can’t attend in person, I want to contribute to this hackathon in three ways:
- do an evaluation of how the reference managers Papers, Mendeley and Zotero (the three reference managers I use) support citations to the biological databases ENA, PDB and Uniprot and what is missing
- look at existing aggregators of this information (e.g. Identifiers.org) to figure out whether the import process can be simplified
- start work on Zotero web translators for these three databases. Zotero is open source software and the web translators are written in Javascript
Please contact me if you are interested in helping with this, e.g. with a joint virtual hackathon on the 18th (or in person in London or Cambridge on October 15 if that works better).
Together with Ian Mulvany from eLife and others from Papers and Mendeley we have also submitted a proposal for a pre-conference workshop/hackathon for the Force2015 Conference in January to work on this for a broader set of databases, which should for example also include software repositories. One question is how we properly handle the citation of large numbers of datasets (1000s to millions), we could for example allow a range of identifiers in a citation. We also need tools to convert identifiers and links in existing documents to proper references, something that we have also discussed on this blog, and we need to discuss how our bibliographic file formats (e.g. bibtex) support these citation types. I said before that I am a big fan of Citeproc YAML (or JSON, the bibliographic format used by CSL) as bibliographic exchange format, and I know that the PLOS Labs hackathon will also touch on this.
Community
While adding reference manager support for a wider range of citations is the first step, the bigger challenge is community support. I don’t think that it is a big mental jump for an author to use the reference manager to cite a biological database rather than typing in the identifier directly in the text (the hard work is registering the identifier in the first place), but this needs support by the community, and in particular journal editors. The important message is that citations should be done in a consistent way and authors don’t have to think about doing this differently for datasets or other relevant resources, or different publishers implementing this differently. I think the paper by Kafkas et al. (2013) clearly shows that our current recommendations for adding identifiers to biological databases is broken, and that we need to do something if we take data citation seriously.
There are several concerns about adding every citation to the references list. One of them is that we shouldn’t mix citations of scholarly articles with citations of other things, e.g. research data. I would argue that not only are we seeing an increasing number of citations to other resources in reference lists, but that we can of course group citations by citation type, in addition to the sorting by appearance in the text or last name of first author that is common now.
Another concern is that citations of datasets are something else that citations to scholarly articles, because the former are typically citations of content created by the same group of people at the time the journal article was also created. I would argue that again we can highlight this by how we display the references, and that I hope that this changes once data citation becomes more widespread.
What should or should not be cited in a scholarly document is of course a big discussion topic. What I am arguing is that everything that is cited should go into the references list, but that doesn’t change at all what should be cited. Personal communications are an example of something that should probably not be cited and therefore should also not go into the references list.
References
Fenner M. Please keep it simple. Published online September 16, 2014. doi:10.53731/r294649-6f79289-8cw05
Kafkas Ş, Kim JH, McEntyre JR. Database Citation in Full Text Biomedical Articles. Larivière V, ed. PLoS ONE. 2013;8(5):e63184. doi:10.1371/journal.pone.0063184
Data Citation Synthesis Group. Joint Declaration of Data Citation Principles. Force11; 2014. doi:10.25490/A97F-EGYK
Fenner M. Citations in Markdown Part 3. Published online June 24, 2013. doi:10.53731/r294649-6f79289-8cw19
Fenner M. Citeproc YAML for bibliographies. Published online July 30, 2013. doi:10.53731/r294649-6f79289-8cw12
Yang S, Han R, Ding J, Song Y. The distribution of Web citations. Information Processing & Management. 2012;48(4):779-790. doi:10.1016/j.ipm.2011.10.002


