This blog post is a follow-up to a post in September (Fenner 2022a), where I announced that I had started working on an archive for scholarly blog posts based on the InvenioRDM open-source repository software. In the last two months, I focussed on two activities – besides lots of physical therapy and other training following a stroke earlier this year (Fenner 2022b): helping to make it easier (and safer) to run InvenioRDM in Docker container infrastructure, and working on converting the bolognese metadata conversion Ruby gem (Fenner 2017) to Python (work in progress on GitHub) to enhance InvenioRDM functionality.
Building an archive of scholarly blog posts faces the same fundamental challenges as repositories for other types of scholarly content, whether data, software, preprints, or journal articles. You have to collect metadata and content, and that approach only scales with standardization and open licenses.
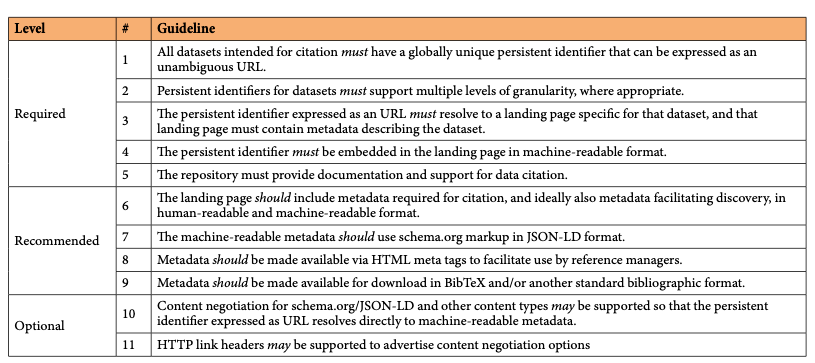
Luckily we already know a lot about required and optional but desired scholarly metadata, and they are fundamentally not different for scholarly blog posts. This means we can take similar approaches as we have for example taken for research data:
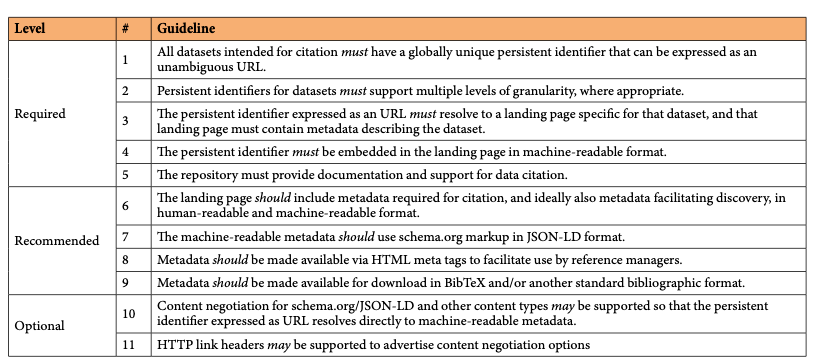
Persistent identifiers for blog posts can be DOIs, as this blog is doing since earlier this year (Fenner 2022). The main advantage of using DOIs is registering standard metadata stored independently of the blogging platform, in case the platform changes or disappears (as has happened several times in the 15 years this blog exists). While there are several blogs using DOIs for their posts, they often fail in guideline #4: the persistent identifier must be embedded in the landing page in machine-readable format. This is important so that reference managers can capture the DOI and retrieve the associated metadata.
When the metadata are embedded directly in the blog post, schema.org markup in JSON-LD format (guideline #7) is much more convenient than HTML meta tags (guideline #8), but for the time being reference managers only work with the latter. The blogging platform used for this blog (https://ghost.org/) has schema.org metadata built in, and there was only a small amount of work needed to expose all metadata needed (or desired) for DOI registration:
- Canonical URL: the DOI for the blog post
- License: the Creative Commons license for the content (this blog uses the Creative Commons Attribution 4.0 License)
- ISSN: the Internal Standard Serial Number of this blog (2749-9952)
An issue I have seen with schema.org metadata is that sometimes they are added by a script running in the browser instead of coming from the server, and this makes metadata harvesting unreliable. Multiple versions and different levels of granularity – a major challenge when working with data and software – luckily is not a major issue with scholarly blogs, in this regard, they behave similarly to preprints and journal articles.
Infrastructure for Archiving Scientific Blog Posts
There are several possible approaches to building infrastructure for scholarly blog posts, and they all have well-known real-world examples:
- A central repository
A good example is the ArXiv.org e-Print archive, which hosts more than two million preprints in physics, mathematics, computer science, and some other fields, and is doing that for more than 25 years. All content and metadata are registered and stored in a central location (since earlier this year using DOIs), and then distributed elsewhere, often domain-specific resources such as ADS (astrophysics) and InspireHEP (high-energy physics). ArXiv is hosted by Cornell University. - A central archive with content published in many places
PubMed (metadata) and PubMed Central (metadata and content) are the main archives of biomedical and life sciences journal literature, again doing this for more than 25 years. The content with metadata is published in many different places but aggregated in PubMed/PubMed Central. Just like ADS and InspireHEP, PubMed and PubMed Central (and Europe PMC) are the central resources for scientists in the field to discover the relevant literature. PubMed uses the PMID, which can be mapped to the corresponding DOI as persistent identifier. For full-text content not included in PubMed Central, PubMed links out to publisher websites. PubMed is hosted by the National Library of Medicine at the U.S. National Institutes of Health (NIH). NIH is the largest funder in the biomedical and life sciences, and its policies help PubMed Central host content. - A central archive with content published elsewhere
The repository Zenodo is the largest generic repository of scholarly content with more than 1.5 million publications, and a million datasets, software, images, and presentations. Almost all content uses open licenses (either one of the Creative Commons licenses or Open Source Initiative approved licenses for software), facilitating the reuse of the content. Zenodo issues DOIs for its content, the DOI points to the Zenodo repository also for content originally registered elsewhere (e.g. software hosted on GitHub). Zenodo is particularly relevant for the planned blog posts archive, as the InvenioRDM software is based on Zenodo software and work is in progress for InvenioRDM to power Zenodo. Zenodo is hosted by CERN, the European Organization of Nuclear Research, the central resource for high-energy physics research.
Based on the above, what makes sense for a scholarly blog post archive?
- Blogs are very decentralized based on their technology and 20-year history. While blogging platforms also have a long history (and Wordpress is the elephant in the room powering more than 40% of all websites), a central blogging platform for science blogs similar to what ArXiv is doing in several fields is neither realistic nor desirable.
- DOIs are a good fit as persistent identifiers for scholarly blogs. A lot of tools and services exist for them (including the InvenioRDM open source software), and the required and desired metadata for blogs are basically covered by DOI metadata (at least for Crossref and DataCite DOIs). Minor issues are that there is no dedicated content type for blog posts and that feature image metadata (supported by schema.org) would be beneficial.
- The business models for DOI registrations need to be adapted to work better for scholarly blogs. A high fixed annual fee (DataCite) or a DOI pointing to a central archive instead of the original content (software in Zenodo) are hurdles for the long tail of independent science bloggers.
- We need business models for sustainable science blogging infrastructure. Advertisements aren't working (the German platform scienceblogs.de is for example closing at the end of year) and individual readers paying for content might work for popular newsletters but doesn't align with Open Science practices. More work is needed, but one key element is cheap and simple infrastructure.
- Existing blogging software (e.g. Wordpress, Ghost, Hugo, or Jekyll) almost works for scholar blogs. Some minor changes (particularly around the canonical URL/persistent identifier) are required to improve the use in reference managers and archiving services.
- We need a standard archiving format for scholarly blogs. JATS (Journal Article Tag Suite) is a standard for scholarly articles, but probably too heavy for blog posts. More work is needed and it should align with RSS (Really Simple Syndication), the 20-year-old standard for distributing content from blogs and similar sources. The biggest gap is maybe a standard way to describe links and references.
- We need aggregation of science blog metadata and content in a central archive. This enables much easier discovery and long-term archiving, I still enjoy reading an interview I did with Geoff Bilder in 2009 (Fenner 2009), and the points he makes are still relevant. The blog posts had moved at least three times over the years and would have greatly benefitted from a DOI, standard archiving format, and a long-term archive as home.
- I have started the work of building the infrastructure for archiving scholarly blogs, but I am fully aware that this is not only a technical challenge but even more so one of governance and community engagement. This needs much more work in 2023 and onwards, and something I look forward to working on jointly with others.
References
Fenner, M. (2022a). Starting Work on the Front Matter Archive [Blog]. Front Matter. https://doi.org/10.53731/9z6rz5d-djbay0y
Fenner, M. (2022b). I spent the last five months in the hospital [Blog]. Front Matter. https://doi.org/10.53731/bkkzj8g-gd14mb6
Fenner, M. (2017). Bolognese: A Ruby library for conversion of DOI Metadata. DataCite. https://doi.org/10.5438/N138-Z3MK
Fenner, M., Crosas, M., Grethe, J. S., Kennedy, D., Hermjakob, H., Rocca-Serra, P., Durand, G., Berjon, R., Karcher, S., Martone, M., & Clark, T. (2019). A data citation roadmap for scholarly data repositories. Scientific Data, 6(1), Article. https://doi.org/10.1038/s41597-019-0031-8
Fenner, M. (2022c). DOI Registrations for all Ghost Blogs [Blog]. Front Matter. https://doi.org/10.53731/fezg09h-hgn1gzm
Fenner, M. (2009). Interview with Geoffrey Bilder [Blog]. Front Matter. https://doi.org/10.53731/r294649-6f79289-8cw1h